Documentation
Comprehensive guides and documentation for all Geinforce Technology tools and services.
Getting Started
Welcome to Geinforce Technology's documentation. This guide will help you get started with our suite of computational tools for drug discovery.
Account Setup
Create an account and get started with Geinforce Technology's platform.
- Visit our registration page
- Enter your details and verify your email
- Explore available tools and plans
- Start using our computational tools
Quick Start Guide
Jump right in with our most popular tools:
- ForceADME - ADME property prediction
- GeinDock Suite - Molecular docking
- ForcePhrase - Natural language processing
- AI Detection - AI-generated content detection
Tools Overview
Geinforce Technology offers a comprehensive suite of computational tools designed to accelerate drug discovery research. Each tool is built on cutting-edge algorithms and trained on extensive datasets to provide accurate and reliable results.
ForceADME
ForceADME is our advanced ADME property prediction tool that helps researchers evaluate the drug-likeness of small molecules. It provides comprehensive analysis of Absorption, Distribution, Metabolism, and Excretion properties.
Key Features
- Comprehensive Property Prediction: Calculate over 30 different molecular and ADME properties
- Multiple LogP Calculation Methods: Including consensus LogP, XLogP3, WLogP, MLogP, and more
- Water Solubility Estimation: Using ESOL, Ali, and SILICOS-IT methods
- Pharmacokinetics Predictions: GI absorption, BBB permeation, P-gp substrate, CYP inhibition
- Drug-likeness Analysis: Lipinski, Ghose, Veber, Egan, and Muegge rule evaluations
- Medicinal Chemistry Alerts: PAINS and Brenk structural alerts
- Interactive Visualization: Including Boiled Egg plot for absorption and BBB permeation
Technology
ForceADME uses a combination of physics-based methods and machine learning approaches:
- RDKit-based molecular property calculations
- Consensus approaches for improved accuracy
- Deep neural networks for complex property predictions
- QSPR (Quantitative Structure-Property Relationship) models
Getting Started with ForceADME
Follow these steps to analyze a molecule using ForceADME:
-
Access ForceADME: Navigate to the ForceADME tool in your dashboard.
-
Input Your Molecule: You can input your molecule in several ways:
- Draw the molecule using the built-in JSME editor
- Enter the SMILES string directly
- Upload a file containing the structure (SDF, MOL)
-
Run Analysis: Click the "Analyze" button to start the computation.
-
View Results: Results will be displayed in an interactive dashboard with detailed sections for each property type.
-
Export Results: You can export the results in various formats including PDF, Excel, and JSON.
Example Use Cases
Example 1: Analyzing Aspirin
SMILES: CC(=O)OC1=CC=CC=C1C(=O)O
Key predicted properties:
- Molecular Weight: 180.16 g/mol
- LogP: 1.31
- Topological Polar Surface Area: 63.60 Ų
- H-Bond Donors: 1
- H-Bond Acceptors: 4
- GI Absorption: High
- BBB Permeant: Yes
- Bioavailability Score: 0.85
Interpretation: Aspirin passes all drug-likeness rules and has favorable ADME properties, which aligns with its well-known pharmacokinetic profile.
Example 2: Comparing Drug Candidates
ForceADME can be used to compare multiple drug candidates to identify the most promising lead compounds:
- Analyze each candidate molecule
- Compare key ADME parameters
- Identify compounds with the best overall profile
- Export comparison reports for team discussion
This approach helps prioritize the most promising candidates early in the development process, saving time and resources.
Input Parameters
| Parameter | Description | Required | Default |
|---|---|---|---|
| SMILES | SMILES string representation of the molecule | Yes | None |
| Molecule Name | Custom name for the molecule | No | "Unnamed Compound" |
| Calculation Mode | Standard or Advanced mode for property calculation | No | Standard |
Output Parameters
ForceADME provides a comprehensive set of calculated properties, including:
Molecular Properties
- Molecular Formula
- Molecular Weight
- Heavy Atom Count
- Aromatic Heavy Atoms
- Fraction Csp3
- Rotatable Bonds
- H-Bond Acceptors
- H-Bond Donors
- Molar Refractivity
- TPSA
Pharmacokinetics
- GI Absorption
- BBB Permeant
- P-gp Substrate
- CYP1A2/2C19/2C9/2D6/3A4 Inhibition
- Log Kp (skin permeation)
Drug Likeness
- Lipinski Rule of Five
- Ghose Filter
- Veber Rules
- Egan (BBB) Rule
- Bioavailability Score
GeinDock Suite
GeinDock Suite provides advanced molecular docking capabilities for predicting binding affinities and interactions between small molecules and protein targets.
Features Overview
- Flexible docking engine with multiple scoring functions
- Support for diverse protein targets and ligand types
- Automated binding site detection
- Batch processing capabilities
- Interactive 3D visualization of docking results
- Integrated with protein preparation tools
- Customizable docking parameters
- Comprehensive binding interaction analysis
- Export results in various formats
- Access to curated protein structure database
How It Works
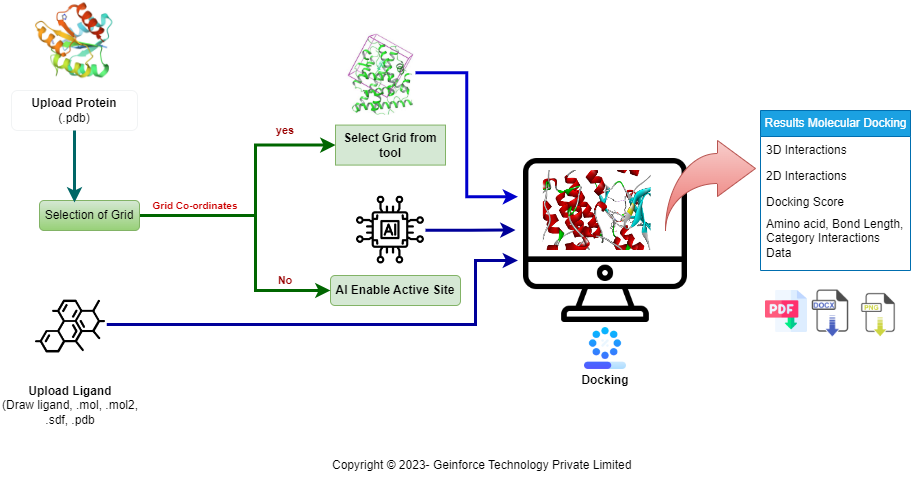
For complete usage instructions, examples, and API documentation, please refer to the GeinDock User Manual.
ForcePhrase
ForcePhrase is our natural language processing tool designed specifically for extracting insights from scientific literature and patents related to drug discovery.
Key Capabilities
Smart Literature Search
Semantic search capabilities across scientific databases, patents, and publications.
Relationship Extraction
Identify compound-target, drug-disease, and other biomedical relationships.
Knowledge Graphs
Build and visualize complex biomedical knowledge networks from literature.
AI Detection
Our AI Detection tool helps identify content that was likely generated by artificial intelligence systems, helping maintain integrity in scientific publications and other written content.
Key Features
- High accuracy detection of AI-generated text
- Support for multiple languages
- Analyzes writing style, patterns, and linguistic markers
- Provides confidence scores and detailed analysis
- Continuously updated to detect output from latest AI models
How to Use
- Navigate to the AI Detection tool
- Input the text you want to analyze (paste directly or upload a file)
- Click "Analyze" to process the text
- Review the results, including the AI probability score and analysis
Example Analysis
Here's an example of how our AI Detection tool analyzes text:
Analysis Results
Verdict: Likely human-written
Confidence Factors
- Specific scientific terminology
- Natural sentence variation
- Domain-appropriate phrasing
- Presence of nuanced qualifiers
GeinBioPredictor
GeinBiopredictor is our tool for bioactivity prediction, designed to estimate the activity of compounds against specific targets using advanced machine learning algorithms.
Features
- Predict bioactivity against hundreds of protein targets
- Estimate IC50, EC50, Ki, and Kd values
- Identify potential off-target effects
- Analyze structure-activity relationships
- Generate bioactivity profiles for lead optimization
API Reference
Geinforce Technology provides comprehensive APIs for integrating our tools into your workflows and applications. This section covers the basics of our API usage.
Authentication
All API requests must be authenticated using API keys. To get your API key:
- Log in to your Geinforce account
- Navigate to Account Settings > API Keys
- Generate a new API key
curl -X GET "https://api.geinforce.com/v1/adme/analyze" \ -H "Authorization: Bearer YOUR_API_KEY" \ -H "Content-Type: application/json"
ADME API
The ADME API allows you to programmatically analyze compounds for their ADME properties.
/v1/adme/analyze
Analyze a compound for ADME properties.
Request Parameters
smiles (string, required): SMILES string of the molecule
name (string, optional): Custom name for the molecule
mode (string, optional): Calculation mode ("standard" or "advanced")
Example Request
{
"smiles": "CC(=O)OC1=CC=CC=C1C(=O)O",
"name": "Aspirin",
"mode": "advanced"
}
Example Response
{
"id": "a1b2c3d4-e5f6-7g8h-9i0j-klmnopqrstuv",
"name": "Aspirin",
"smiles": "CC(=O)OC1=CC=CC=C1C(=O)O",
"properties": {
"molecular_formula": "C9H8O4",
"molecular_weight": 180.16,
"logp": 1.31,
// Additional properties...
}
}
Docking API
The Docking API provides programmatic access to our molecular docking engine.
SDKs & Libraries
Geinforce provides software development kits (SDKs) in several popular programming languages to make integration with our APIs easier.
Python SDK
Our Python SDK is ideal for data scientists and computational chemists working in Python environments.
pip install geinforce-sdk
R Package
Our R package provides easy access to Geinforce APIs from R statistical environment.
install.packages("geinforceR")
Tutorials
Learn how to use Geinforce Technology's tools effectively with our step-by-step tutorials.
Getting Started with ForceADME
20 minLearn the basics of using ForceADME for ADME property prediction.
Molecular Docking with GeinDock
30 minSet up and run your first molecular docking experiment.
API Integration for High-Throughput Screening
45 minAutomate your compound screening workflow using our APIs.
Data Visualization and Reporting
25 minCreate beautiful and informative reports from your analysis results.
Frequently Asked Questions
Troubleshooting
Encountering issues? Here are solutions to common problems.
Common Issues & Solutions
Invalid SMILES Error
Problem: Receiving an "Invalid SMILES string" error when analyzing a molecule.
Solution: Check for syntax errors in your SMILES string. Make sure valences are correct and all atoms are properly defined. You can validate your SMILES using our SMILES validator tool or third-party services.
API Authentication Failed
Problem: Receiving a 401 Unauthorized error when making API calls.
Solution: Verify that you're using the correct API key and that it's active. Make sure you're including the Authorization header correctly. Check if your subscription plan is active and has sufficient API credits remaining.
Docking Job Fails
Problem: GeinDock job fails with an error.
Solution: Check that your protein structure is complete and properly prepared. If using a custom ligand, verify its structure. For large jobs, try splitting into smaller batches. If issues persist, contact support with your job ID for assistance.
Need Additional Help?
If you can't find a solution to your problem, please contact our support team. Include detailed information about the issue you're experiencing for faster resolution.
Release Notes
Stay up to date with the latest updates and improvements to our platform.
Version 2.8.0 - June 2024
New Features
- Added support for macrocycle ADME prediction in ForceADME
- Introduced advanced binding site prediction in GeinDock
- Released beta version of ForcePhrase for literature mining
- Added batch processing for AI Detection tool
Improvements
- Enhanced LogP prediction accuracy with new consensus algorithm
- Improved GeinDock scoring function for fragment-like compounds
- Optimized API performance with 2x throughput increase
- Updated user interface for better mobile experience
Bug Fixes
- Fixed rare issue with aromatic ring detection in certain heterocycles
- Resolved API timeout on large batch submissions
- Fixed sorting in analysis history view
- Corrected minor display issues in Safari browser
Version 2.7.2 - April 2024
Improvements
- Updated molecular visualization engine for better performance
- Added 200+ new protein structures to the docking database
- Enhanced report generation with new visualization options
Bug Fixes
- Fixed issue with export to Excel on certain property sets
- Resolved authentication issue for some API clients
- Fixed inconsistent behavior in JSME molecular editor